CIRM's goal in establishing the CESCG is to apply genomics and bioinformatics approaches to stem cell research to accelerate fundamental understanding of human biology and disease mechanisms, enhance cell and tissue production and advance personalized cellular therapeutics.
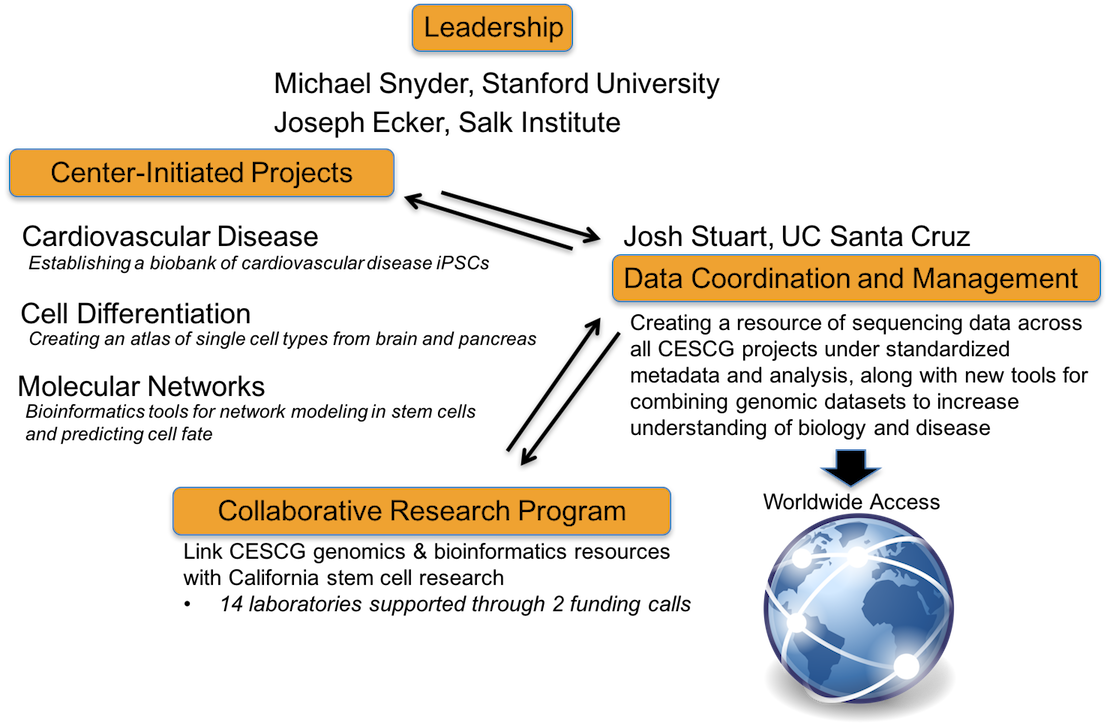
The CESCG is composed of Operational Cores at Stanford University and at the Salk Institute and a Data Coordination and Management Core at the University of California Santa Cruz, which currently support the following research programs:
Created in 2014 through a $40 million award by California's stem cell agency, the California Institute for Regenerative Medicine (CIRM), the Center of Excellence in Stem Cell Genomics (CESCG) is spearheading the investigation into how stem cells can be used to treat disease.
This Center merges the genomics expertise of the Stanford University research team under the seasoned leadership of Mike Snyder, with the cutting edge expertise of the Salk Institute and University of California, San Diego groups in epigenomics, led by Joseph Ecker. These two leaders in the field of genomics have extensive experience working on large-scale projects and have been working in national and international consortia for generating and distributing genomics data. This Center of Excellence also includes the experience of Josh Stuart and David Haussler, leaders in data coordination and management, from the University of California, Santa Cruz (UCSC), extending collaborative relationships that have been developed over the past decade.
Our vision is to advance Stem Cell research in the State of California by establishing the Center of Excellence in Stem Cell Genomics (CESCG). The Center is generating banks of data and iPS cell lines that are valuable resources to the entire CIRM community and provide important insights into stem cell research. It also serves as an important focal point for collaborative projects with other CIRM investigators and makes genomics capabilities available to the entire regenerative medicine community.
The center will have state-of-the-art Center-Initiated Projects (CIPs) whose data and iPS cell lines will serve as a valuable resource to the entire CIRM community and provide important insights into stem cell research. It will also serve as an important resource for nucleating collaborative projects with other CIRM investigators and making genomics capabilities available to the entire regenerative medicine community.
This Center-Initiated Project is studying matched set of iPSC lines from patients with cardiovascular disease and the corresponding whole genome sequences and genome-wide expression analyses. The primary goal of this CIP is to establish a biobank of extensively well-characterized iPSC lines and to validate the utility of this resource by modeling two highly prevalent familial forms of cardiovascular disease, DCM and HCM, and performing drug screening studies. This CIP will also investigate genome stability of iPSC lines, which is crucial for understanding their therapeutic value.
This Center-Initiated Project is systematically characterizing the heterogeneous cell subpopulations within normal and pathological tissues of the human brain and pancreas, and determining the gene expression and epigenetic properties of each member of the organ lineage tree, from stem cells to terminally differentiated cells. Using this data, this project will compare the stem cell differentiation hierarchy of normal tissues to that of the disease states to elucidate underlying mechanisms of pathogenesis, and identify gene expression and epigenetic markers for premalignant or malignant stem cell contaminating cell products intended for patient clinical trials.
Cellular differentiation and maintenance of pluripotency involve a complex series of events governed by molecular networks. The overall objective of this Center-Initiated Project is to develop a suite of bioinformatics tools and resources for advanced analysis of -omics data generated by the CIRM Genome Center, with the goals of formulating molecular network models and guiding predictions of cell fate.
Michael Snyder, PhD
Michael Snyder (PI) is
Professor & Chair of Genetics at Stanford University and Director of the
Stanford Center for Genomics &
Personalized Medicine. He is an expert in the field of functional genomics and
proteomics, and he has developed many genomic technologies and informatics pipelines.
Joseph C. Wu, MD, PhD
Joseph C. Wu (co-PI) is
Professor of Medicine/Cardiology and Director of the Stanford Cardiovascular Institute.
He is an expert in cardiac developmental biology and in iPSC drug screening platforms.
Kristin K. Baldwin, PhD
Kristin K. Baldwin (co-PI)
is an Associate Professor at the Scripps Research Institute. Her laboratory generated
the first iPSC lines that could produce an entire organism.
David Haussler, PhD
David
Haussler (co-PI) is a Distinguished Professor of Biomolecular Engineering at
the University of California, Santa Cruz and Investigator at the Howard Hughes Medical
Institute. He is the world's expert with human genome analysis and data management.
Carlos D. Bustamante, PhD
Carlos
D. Bustamante (collaborator) is Professor of Genetics at Stanford University and
Director of the Stanford Center
for Computational, Evolutionary and Human Genomics.
He is a population geneticist and has received the MacArthur Fellow Foundation Award
(a.k.a. the "Genius Award") for his research in population genetics for humans,
animals, and plants.
Stephen Quake, DPhil
Stephen Quake (PI) is the
Lee Otterson Professor of Bioengineering and Applied Physics at Stanford University,
and an Investigator at the Howard Hughes Medical Institute. He is recognized as one of
the fathers of microfluidics and a pioneer of genomics. He has received many prizes for
his discoveries and inventions, including the Lemelson-MIT Prize for invention and
innovation, the Nakasone Prize from the Human Frontiers of Science Foundation, the
Sackler International Prize for Biophysics, and the Promega Biotechnology Award from
the American Society for Microbiology.
Michael F. Clarke, MD
Michael F. Clarke (co-PI) is the Karel and Avice Beekhuis Professor
in Cancer Biology at Stanford University and the Associate Director of the
Stanford Institute
for Stem Cell Biology and Regenerative Medicine.
Trey Ideker, PhD
Trey Ideker (PI) is Professor in the University of California
San Diego, Department of Medicine. His lab was named one of the Top Ten Innovators of
2006 by Technology Review magazine and he was the 2009 recipient of the Overton Prize
from the International Society for Computational Biology.
Josh Stuart, PhD
Josh Stuart (co-PI) is
Professor in the Department of Biomolecular Engineering at the University of California,
Santa Cruz. Since 2009, he has co-directed a genome data analysis center for the Cancer
Genome Atlas (TCGA) project and leads the TCGA's pan-cancer project to investigate
patterns across tumor types. He plays a key role in several national and international
consortia including TCGA, Stand Up To Cancer, and the International Cancer Genomics
Consortium.
Mark D. Adams, PhD
Mark D. Adams (co-PI) is the Scientific Director of the J. Craig Venter Institute.
Through 13 years working at NIH, The Institute for Genomic Research (TIGR), and Celera
Genomics, he was responsible for applying new sequencing technologies and analytical
approaches to sequencing of ESTs, microbial genomes, and, ultimately, the human genome.
Richard Scheuermann, PhD
Richard
Scheuermann (co-PI) is the Director of Informatics at the J. Craig Venter Institute.
He serves as one of the coordinating editors of the Open Biomedical Ontology (OBO)
Foundry, and has been a contributing developer of both the Cell Ontology and the
Ontology of Biomedical Investigations. He has also served on the Scientific Advisory
Board of the Gene Ontology.
A major part of the CESCG mission is to establish a Collaborative Research Program (CRP) to support the genomics research needs of stem cell investigators in California. Through the CRP funding of individual collaborative research projects, the CESCG will provide expertise and resources for the development and application of new and innovative genomic and epigenomic approaches for human stem cell biology and regenerative medicine. The objective is to combine stem cell resources with CESCG genomic and bioinformatics approaches to accelerate fundamental understanding of human biology and disease mechanisms, enhance cell and tissue production and advance personalized cellular therapeutics.
Data Release Policy
All genomics data generated in Collaborative Research Program projects will be made available to the research community through the Data Coordination and Management (DC&M) Center of the CESCG no later than the time of publication. We encourage early data release prior to publication.
Data Access Policy
Genomics data collected under the CESCG Collaborative Research Program will be stored at the CESCG DC&M Center, and made available to researchers worldwide. Access to the genomics data will be provided to all qualified researchers.
To access the genomics data, users will need to create an account by:
Genomics/epigenomics Services Available
CESCG Sequencing and Informatics Services (updated 10/29/14)
(The informatics costs are to be applied only to the CESCG portion of Comprehensive budgets.)
Crooks (UCLA)
Pluripotent stem cells (PSC) are an attractive source for deriving hematopoietic stem cells (HSC) for the treatment of blood disorders. However, yet poorly understood molecular blocks have prevented the generation of functional HSCs in culture. Preliminary studies imply that although cells that possess the immunophenotype of human HSC can be generated, the transcriptome of these cells is dysregulated at multiple levels, including protein coding genes, non-coding RNAs, and mRNA splicing. The Crooks lab will now create a comprehensive transcriptome and epigenome map of human HSC ontogeny by comparing human hematopoietic tissues and embryonic stem cell (ESC)-derived cells, providing a data set in which a limited number of differentially regulated RNAs likely to have functional impact can be identified. Importantly, this analysis will pinpoint key defects that underlie the poor function of ESC-derived hematopoietic cells, and offer new solutions for overcoming these molecular barriers. Moreover, as the hematopoietic hierarchy offers a powerful system for dissecting how transcriptome changes govern tissue development and homeostasis, these studies will provide broader insights to the regulation of stem cell fate decisions and how to induce proper developmental programs for generating tissue stem cells for regenerative medicine.
Geschwind (UCLA)
Advances in genetics and genomics over the last decade have led to the identification of specific genetic variants that account for approximately 20% of the risk for autism spectrum disorder (ASD). Many of these variants represent rare, large effect size mutations. The Geschwind lab has demonstrated a convergent pattern of disrupted gene expression in post mortem ASD brain, but it is not known how multiple distinct mutations lead to this convergent molecular pathology. To address this question, the lab will perform large-scale genomic and phenotypic characterization of induced pluripotent stem cell (iPSC)-based models of ASD, representing patients with distinct ASD risk mutations, patients with idiopathic ASD and healthy controls. Neurons will be generated from iPSC using an innovative 3D neural differentiation system that generates a laminated human cortex, including synaptically connected neurons and astrocytes. These in vitro models will be phenotyped for morphological and physiological abnormalities using automated assays and entire transcriptional networks will be analyzed during in vitro development. In addition, whole genome sequence will be obtained and together these data will be used to identify potential causal factors and key regulatory drivers of the disease. This will provide new mechanistic insight in ASD pathophysiology and provide an invaluable and unprecedented resource for the field. Furthermore, this project leverages iPSC lines generated as part of the CIRM hiPSC Initiative and capitalizes on tools being developed by the Ideker lab as part of the broader CESCG toolbox.
Kriegstein (UCSF)
Cortical progenitor cells give rise to the diverse populations of cortical neurons. Many protocols have been developed to generate cortical neurons from pluripotent stem cells, including several recent protocols that use aggregate culture methods to differentiate human pluripotent stem cells into cerebral organoids. Cerebral organoids consist of heterogeneous populations of progenitors and neurons but the extent to which these in vitro-derived cell types resemble their endogenous counterparts remains unknown. To address this issue, the Kriegstein lab will use single cell RNA profiling at multiple stages of cerebral organoid differentiation and compare these in vitro gene expression profiles with data they are generating for primary human cortical tissues. These comparisons will provide objective measures of the efficiency and accuracy of various differentiation protocols. The ultimate goal of this analysis is to improve differentiation protocols, a necessary effort in order to realize the potential of stem cells for cell replacement therapy as well as for disease modeling.
Bruneau (UCSF)
The goal of this project is to develop an integrated understanding of epigenomic regulation of human cardiac differentiation, in order to apply this knowledge to congenital heart disease (CHD). The Bruneau lab will use patient-specific and engineered human induced pluripotent stem cell (hiPSC) lines to evaluate the impact of heterozygous missense mutations in CHD-associated transcriptional regulators on gene expression and a broad range of chromatin states in cardiac precursors and cardiomyocytes. In support of these goals, the lab will develop human cellular models that represent the two major cardiac cell types, atrial and ventricular cardiomyocytes, using hiPSC engineered to allow the isolation of these two cell types. And finally, they will define dynamic 3D maps of genomic interaction in human cardiac differentiation. This will yield a complete view of the effects of mutations in key chromatin modifying genes on gene regulatory causes of CHD, and an unprecedented view of human cardiac lineage commitment.
Frazer (UCSD)
Sudden cardiac arrest (SCA) is a leading cause of death among adults over the age of 40 in the United States. It is usually caused by ventricular arrhythmias (irregular heartbeats) due to abnormalities in the heart's electrical system. In addition to naturally occurring SCA, drug therapies can cause a form of acquired arrhythmia that leads to SCA. Clinical risk factors for drug-induced arrhythmias include gender, age and existent cardiac and/or liver disease, and there is evidence for a genetic predisposition. The Frazer lab has generated human induced pluripotent stem cells (hiPSCs) from 225 individuals and has established protocols for large-scale derivation of human cardiomyocyte lines from these hiPSCs. They will use these lines for the study of ventricular arrhythmias, both in the naïve state or triggered by drug administration. The lab has NIH funding to identify genetic variants that are associated with electrophysiological phenotypes, while this work will identify genetic variants associated with genome-wide RNA expression levels and the epigenome. Together, these data will provide new knowledge of the biology of arrhythmias that may be exploited to improve treatment options for SCA.
Fan (UCLA)
Human overgrowth syndrome is a class of complex genetic disorders characterized by systemic or regional excess growth compared to peers of the same age. Overgrowth individuals with mutations in DNMT3A, a de novo DNA methyltransferase, are significantly taller, have distinctive cranio-facial features, and exhibit intellectual disability. In this study, the Fan lab will precisely map transcriptome and epigenome dynamics during development of disease-relevant tissues derived from embryonic stem cells that contain a spectrum of DNMT3A knock-in point mutations. The goal is to identify convergent target genes and signaling pathways perturbed in overgrowth syndrome, leading to a rich resource and novel approach to understanding the mechanisms of this disease.
Sanford (UCSC)
Alternative pre-mRNA splicing contributes to the regulation of gene expression and protein diversity. Many human diseases, including Amyotrophic Lateral Sclerosis, Frontotemporal Lobar Degeneration and cancer, are caused or exacerbated by aberrant RNA processing. The goals of this study are to investigate RNA processing during neuronal differentiation of human pluripotent stem cells, using innovative genomics approaches. The knowledge gained is not only critical for understanding how splicing is regulated to control differentiation but also for discovering how genetic variants such as inherited disease mutations disrupt gene expression and function.
Belmonte (Salk)
Somatic cells can be reprogrammed to an induced pluripotent stem cell (iPSC) state by transient forced expression of the 4 Yamanaka factors (4F) (Oct4, Sox2, Klf4, cMyc). This process has been widely explored in vitro to generate a variety of cell types or to rejuvenate them for their therapeutic application by autologous transplantation. However, transplantation is an invasive procedure and challenged by access to the niche, retention and integration of the graft in addition to the safety and functionality concerns of in vitro-derived cells. An alternative provocative approach is to reprogram cells in vivo. Nevertheless, the effect of 4F in vivo is largely unknown at the molecular level. The major goal of this proposal is to characterize the molecular dynamics of cells undergoing 4F-induced reprogramming in vivo in the mouse. Specifically, we are aiming to identify the changes in transcriptome, DNA methylome and histone methylation status. Toward this end, we will make use of a transgenic mouse that carries a Doxycycline-inducible cassette of 4F whose activity will be also conditional upon Cre expression to control the timing and location of the reprogramming in vivo. The dynamics of several cell types of different embryonic origin will be analyzed through time at the populational and single cell levels. We will also characterize the behavior of the cells undergoing a short-term induction followed by a recovery phase to evaluate if the partially reprogrammed cells will return to their original molecular phenotype or adopt an intermediate, de-differentiated phase. One potential therapeutic application of this approach is the rejuvenation of old cells. Thus, we will compare aged samples exposed to partial reprogramming with the young samples to evaluate if partial reprogramming confers a youthful molecular signature to old cells. Elucidation of the molecular dynamics of in vivo reprogramming in the mouse in collaboration with CESCG will enhance our understanding of the possibilities and risk factors of using this technology in regenerative medicine.
Chi (UCSD)
The overall goal of this proposal is to discover gene regulatory networks that will facilitate the differentiation of human pluripotent stem cells (hPSC) into ventricular and atrial cardiomyocyte (CM) lineages for regenerative therapies as well as for more precise modeling and treatment of human congenital or adult heart disease. Toward this goal, in Aim 1, we will perform RNA-seq, ChiP-seq for five histone modifications, and Hi-C analyses on extracts from human CMs purified from each of the four cardiac chambers of human hearts, [right ventricle (RV), left ventricle (LV), right atrium (RA), and left atrium (LA)] to define the transcriptional profiles and genomic regulatory networks for these in vivo specific CM lineages. In Aim 2, a comparable genomic dataset will be obtained for in vitro hPSC-derived ventricular and atrial CMs, at stages selected to correspond to those of characterized human heart samples. Stated objectives of this CESCG Call 2 are to combine genomic and bioinformatics approaches with stem cell research to accelerate fundamental understanding of human biology and disease mechanisms, enhance cell and tissue production, and advance personalized cellular therapeutics. Thus, these cardiac gene expression profiles and epigenetic landscapes to identify enhancers and their cognate promoters will provide critical insights into gene regulatory networks which can be used to identify individual myocardial states and further manipulated to generate specific and mature hPSC-CM lineages that may better recapitulate their in vivo CM counterparts for regenerative therapies and disease modeling. Additionally, these cardiac enhancer/promoter datasets may also provide insights into the significance of potential disease non-coding genetic variants discovered in whole genomic sequencing and genome-wide association heart studies.Corn (UC Berkeley)
Sickle cell disease (SCD) is a devastating genetic disorder that affects ~100,000 primarily African American individuals in the USA, including 5,100 in California. In SCD, a Glu to Val point mutation in the ß-globin gene renders the resultant sickle hemoglobin prone to polymerize and damage the red blood cell. We have used CRISPR-Cas9 genome editing to develop methods to correct the sickle allele in hematopoietic stem cells (HSCs) and, together with SCD experts at Children's Hospital Oakland, are in the process of developing proof-of-concept for a clinical trial to cure SCD via transplantation of gene-corrected autologous HSCs from patients. Our goal in this CESCG CRP is to establish the efficacy and safety of sickle correction in HSCs via targeted and unbiased sequencing. Together with the CESCG we will use next-generation sequencing to determine the extent of allele conversion in edited HSCs both in vitro and after in vivo engraftment in a mouse model. To establish the safety of editing, we will use three sequencing-based approaches. First, we will use custom amplicon-based resequencing to quantify undesired editing events at related globin genes and at sites computationally predicted as potential off-targets based on sequence similarity. Second, we will use established cancer resequencing panels to uncover low-frequency off-target events at genes known to be involved in tumorigenesis, with a focus on annotated tumor suppressors. Third, we will use unbiased capture and sequencing methods, such as the recently described GUIDE-Seq method, to uncover off-target events in the context of the entire human genome. This approach will be critical in providing key data to move editing SCD allele towards the clinic and will also provide an important precedent for establishing efficacy and safety metrics for therapeutic gene editing in HSCs.Jones (Salk)
The Wnt3a/ß-catenin and Activin/SMAD2,3 signaling pathways synergize to induce hESC/iPSC differentiation to mesoderm, an intermediate step in the development of heart, intestine, liver, pancreatic and other cell lineages. Our lab recently investigated the transcriptional mechanism underlying the Wnt-Activin synergy in human embryonic stem cells using genomewide ChIP-seq, GRO-seq, and 3C studies. We found that Wnt3a signaling stimulates the assembly of ß-catenin:LEF-1 enhancers that contain a unique variant form of RNAPII, which is phosphorylated at the Ser5, but not Ser7, positions in the CTD heptad repeats. Importantly, this variant RNAPII was also found at all hESC enhancers, whereas RNAPII at active promoters was highly phosphorylated at both Ser5 and Ser7. Phosphorylation of RNAPII-Ser7 in hESCs is mediated by the positive transcription elongation factor, P-TEFb (CycT1:CDK9), and we showed that ME gene activation depends on P-TEFb. Interestingly, Wnt3a signaling induces enhancer-promoter looping at mesendermal (ME) differentiation genes, which is facilitated by binding of ß-catenin to cohesins. By contrast, Activin/SMAD2,3 did not affect enhancer-promoter interactions, but instead strongly increased P-TEFb occupancy and RNAPII CTD-Ser7P at ME gene promoters. Moreover, Activin is required for CTD-Ser2P and histone H3K36me3 at the 3' end of ME genes. Lastly, we found that many ME genes, including EOMES and MIXL1, were potently repressed by the Hippo regulator complex, Yap1:TEAD, which selectively inhibits P-TEFb elongation, without affecting SMAD2,3 chromatin binding. Thus Wnt3a/ß-catenin-induced gene looping synergizes with Activin/Smad2,3-dependent elongation to strongly up-regulate ME genes. Our unpublished data show that CRISPR/Cas-mediated loss of Yap1 also promotes hESC differentiation to mesoderm, but not ectoderm, through increased Activin signaling. Here we will characterize these cells to define how Yap1 is recruited to ME genes and disrupts the P-TEFb CTD kinase to control the earliest steps of hESC differentiation. Further studies will analyze how Yap1 switches to cooperate with Wnt3a at later stages in cardiomyocyte development.Loring (Scripps)
We are performing preclinical studies for an autologous cell therapy for Parkinson's disease. We have generated iPSCs from 10 Parkinson's patients for our initial cohort, based on criteria established from earlier studies using fetal tissue. Our clinical partner, Dr. Melissa Houser, is Director of the Movement Disorders Clinic at the Scripps Clinic in La Jolla. Our preliminary data include optimized, robust, and reproducible methods for differentiating different lines of patient-specific iPSCs into dopamine neuron precursors, which have shown efficacy in animal models. These are the cell preparations that we plan to transplant to patients to restore their motor control. We also cite our recently published data on genomic stability of human pluripotent stem cells, which pinpoint genomic aberrations that occur when these cells are cultured for very long (>2 years) periods. We include another of our studies, in press in Nature Communications, in which we performed comprehensive analysis of human iPSCs produced by three different reprogramming methods, using whole genome sequencing and de novo genome mapping methods to show that it is unlikely that reprogramming itself will introduce mutations that compromise the safety of iPSCs for therapy. For this grant application, we request funds to perform whole genome and RNA sequencing as quality control measures to assure that the cells that are used for transplantation have no deleterious mutations and are the correct cell type. The mRNAseq studies will expand on our earlier work, funded by CIRM, in which we developed a genomic diagnostic test to determine whether human cells are pluripotent. This gene expression-based diagnostic test, called PluriTest®, is now the most popular assay for pluripotency, recommended by the NIH and used more than 12,000 times since the website (www.pluritest.org) became active. The genome sequencing studies will improve upon our earlier assessments of genomic stability based on SNP genotyping. These studies will allow us to provide the most rigorous predictive assessments of cell therapy safety and efficacy for clinical stem cell applications, and help to set the standards for future clinical studies using human stem cell-derived products.Yeo (UCSD)
In recent years, the importance of post-transcriptional gene regulation (PTGS) underlying neurodegenerative disorders such as amyotrophic lateral sclerosis (ALS), Alzheimer's and Parkinson's disease has increased tremendously with the growing number of RNA binding proteins (RBPs) found mutated in patients. Specifically, despite the hundreds of mutations found within these RBPs in patients with ALS, we still do not understand (1) how and why they cause motor neuron degeneration while leaving other cell-types in the central nervous system such as glial cells relatively unscathed, and (2) what aspects of PTGS these mutations affect in these cells. Thus this represents an unmet medical need. In order to study whether these disease-associated mutations result in cell-type specific PTGS, we propose to utilize induced pluripotent stem cells (iPSCs) from ALS patients harboring disease-causing mutations and generate mature neurons, a highly relevant cell-type in ALS. RBPs interact with specific sequences or structural features within transcribed RNAs to affect PTGS. In this proposal we will test specific hypotheses that these mutant RBPs affect cell-type specific alternative splicing and sub-cellular mis-localization of target mRNAs, both highly relevant to known normal functions of these RBPs. We will focus on abundantly expressed RBPs (TDP-43, FUS/TLS, hnRNP A2/B1 and Matrin3) that have been implicated in ALS. We already have generated patient-specific iPSC lines with TDP-43, FUS/TLS and hnRNP A2/B1 mutations. We will use highly optimized protocols to differentiate iPSCs to neurons. For the first time, we will identify mutant-dependent sub-cellular mis-localization of alternative isoform mRNAs in neurons as a novel hypothesis for disease pathology in ALS. To address whether these mutations cause cell-type specific aberrations in alternative splicing, we will utilize single-cell RNA-seq analysis to measure alternative splicing in the neuron culture system, which contains the requisite heterogeneous mixture of glial cells to maintain a healthy neuron differentiation and physiology. For the first time, we will identify mutant-dependent cell-type specific alternative splicing as a hypothesis for disease pathogenesis. These stem-cell based mRNA signatures are a critical resource that we will compare to post-mortem patient material to identify potential therapeutic targets.Weissman (Stanford)
The goal of this program is to address a fundamentally unsolved issue in human development and stem-cell differentiation: how does human mesoderm become diversified into an array of therapeutically-relevant tissues, including bone, cartilage, skeletal muscle, heart, kidneys and blood? A comprehensive understanding of how these tissues all differentiate from a common embryonic mesodermal source might enable us to artificially generate these diverse lineages from human embryonic stem cells (hESCs) for regenerative medicine. Human mesoderm development remains a terra incognita, because it unfolds during human embryonic weeks 2-4, when it is ethically impossible to retrieve fetuses for developmental analyses. Because human mesoderm ontogeny has never been systematically described, the identity of key developmental intermediates and the order of lineage transitions remain unclear. A clear "lineage tree" of mesoderm development would expand developmental and cell biology and enhance the therapeutic potential of regenerative medicine. Hence our goal is to chart a comprehensive map of human mesoderm development, including the specifics of blood development (hematopoiesis), and to systematically reconstruct its component lineage intermediates and their progenitor-progeny relationships.